Holobiome: Research I Articles I Updates I News I Series I Feed
Holobiome is a blog series that offers an AI-assisted summary of the latest research articles on human microbiome.
Seeding Health Before Birth: How Maternal Diet Shapes the Infant Microbiome in the FeFiFo‑MOMS Trial
The FeFiFo-MOMS trial is breaking new ground by exploring how dietary choices during pregnancy can shape the maternal and infant microbiome. Modern diets, often low in fiber and fermented foods, have been linked to reduced gut microbial diversity—a factor associated with increased risks of inflammation and metabolic disorders. Pregnancy is a critical period, as the maternal microbiome not only influences maternal health but also seeds the infant’s early microbial community, potentially shaping the child’s immunity and metabolism for years to come. This study specifically examines whether increasing microbiota-accessible carbohydrates from whole-food fiber and incorporating fermented foods rich in live microbes can promote a healthier and more diverse microbial ecosystem during this crucial window.
Conducted between January 2022 and August 2023,the trial recruited 135 healthy pregnant women in California during mid-gestation (12–22 weeks). Participants were randomized into four groups: fiber-focused diets, fermented food–rich diets, a combination of both, or a usual-diet control group. To support adherence, participants received personalized coaching and tracked their diets through a digital platform. The researchers are performing extensive sampling—stool, blood, vaginal swabs, and breast milk from mothers, as well as stool samples from infants—to monitor how the microbiome and immune markers respond to these dietary interventions. The primary aim is to assess whether infants born to mothers in the intervention arms show higher gut microbial diversity at one month of age compared to controls, alongside secondary outcomes like maternal microbiome shifts and reductions in inflammatory markers.
What makes this study significant is its emphasis on whole foods rather than isolated probiotic or prebiotic supplements. By targeting dietary patterns that naturally nurture a broad spectrum of beneficial microbes, FeFiFo-MOMS is testing a sustainable and accessible approach to microbiome modulation. If these interventions prove successful, they could inform new prenatal nutrition guidelines focused not just on maternal health but also on fostering a resilient microbial foundation for infants. The findings could mark a shift toward food-based strategies to enhance microbial diversity, moving beyond capsule-based probiotic solutions to embrace the complexity of the maternal-infant microbiome connection.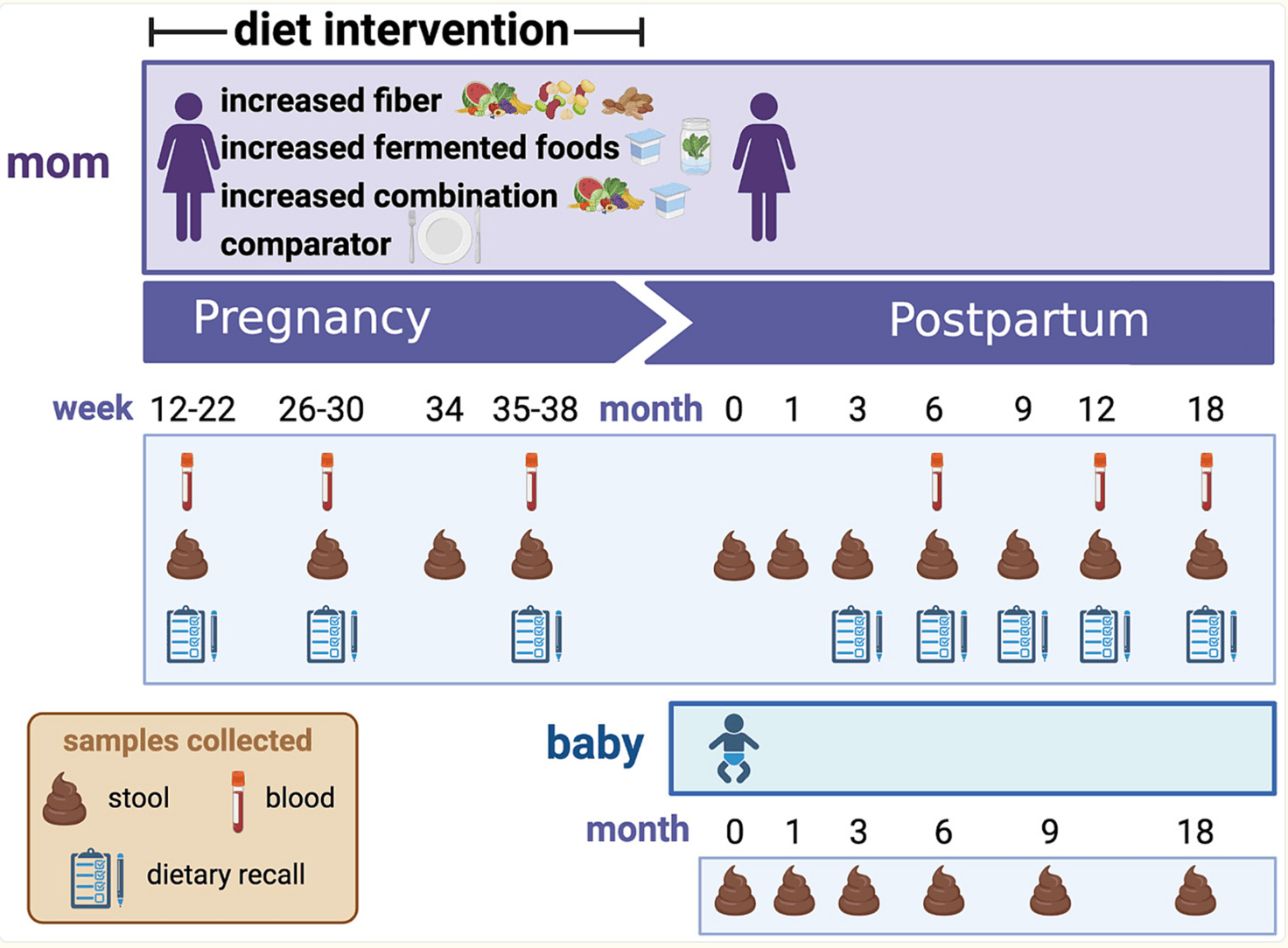
Trial Design
Swallowed but Not Settled: Lessons from a Probiotic Trial on Recurrent Vaginal Infections
Pregnancy places extraordinary pressure on the vaginal ecosystem: hormonal shifts, altered immunity, and the frequent use of antibiotics or antifungals create openings for bacterial vaginosis and vulvovaginal candidiasis to return again and again. A healthy vaginal microbiome is normally dominated by Lactobacillus species that acidify and protect the niche, but treatment-induced disruptions can leave the community in dysbiosis and vulnerable to relapse—risks that carry downstream obstetric consequences for both mother and infant.
This double-blind Israeli trial asked whether a high-dose oral blend of six gut-friendly strains (Bifidobacterium bifidum Bb-06, B. lactis Bi-07, Lactobacillus acidophilus La-14, L. paracasei Lpc-37, L. rhamnosus Lr-32, and Streptococcus thermophilus) could act as a microbiome “booster shot” once an acute infection had been cleared. Forty-seven pregnant women took two capsules per day (≈1.2 × 10¹⁰ CFU) or placebo for roughly four months, with monthly smears and MALDI-TOF screens tracking who recolonised with protective lactobacilli and who slid back into dysbiosis. Despite the ambitious dose, none of the capsule strains were ever recovered from the vagina, and recurrence of any vulvovaginal infection was numerically higher—not lower—in the probiotic arm (67 % vs 48 %; p = 0.19).
For microbiome scientists, the takeaway is clear: oral delivery alone may not bridge the gut-vagina divide. The study highlights formidable ecological barriers—gastric acidity, competitive gut transit, and the cervical mucus plug—that keep most ingested microbes from reaching the lower genital tract. Future strategies will likely need strains with proven vaginal tropism, targeted intravaginal delivery systems, or synbiotic approaches that nourish resident Lactobacillus rather than trying to reseed them from afar. Until then, simply swallowing more probiotics may do little to restore the delicate microbial shield that pregnancy so often disturbs.
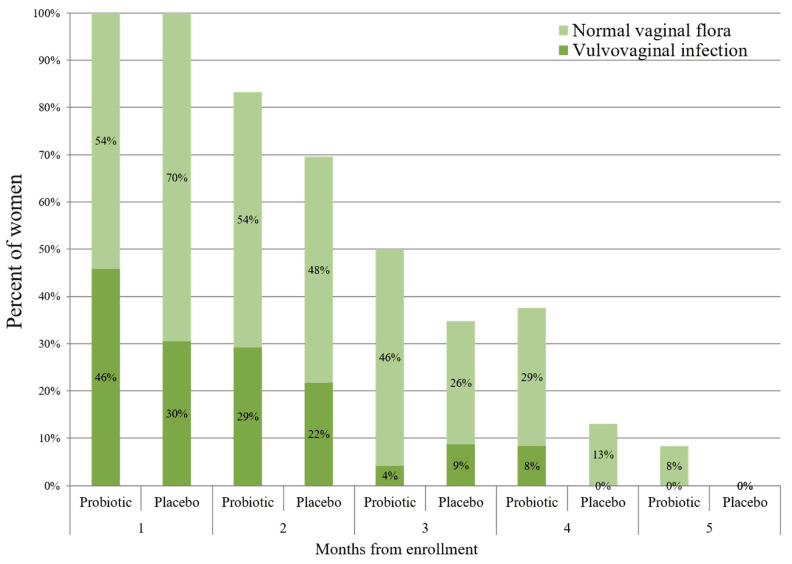
The rate of vulvovaginal infections (VVIs) throughout this study in the probiotic (N = 24) and placebo (N = 23) groups. The study participants were invited for a vaginal swab once in every month until delivery. Repeated vaginal swabs for verifying VVI eradication are not presented. p > 0.05 for all the time points.
Bridging the Microbial Gap: 2′-Fucosyllactose in Infant Formula
Human milk oligosaccharides (HMOs) are the unsung architects of an infant’s first microbial community, selectively feeding Bifidobacterium species that dominate the healthy neonatal gut. Among them, 2′-fucosyllactose (2′-FL) is both abundant and structurally tailor-made to nourish these early colonizers. The new randomized trial in Nutrients asked whether fortifying a standard galacto-/fructo-oligosaccharide (GOS + FOS) formula with 2′-FL could nudge the microbiome of formula-fed babies closer to that of their breast-fed peers—a critical question as many infants worldwide rely on formula from day one.
The investigators followed 87 healthy infants divided into three cohorts: a 2′-FL–supplemented formula group (n = 29), a GOS + FOS control formula group (n = 30), and an exclusively breast-fed reference group (n = 28). Fecal samples collected in the first and fourth months revealed that, after just three months of feeding, Actinobacteriota—driven largely by Bifidobacterium—became the dominant phylum in both the 2′-FL and breast-milk groups (≈60 % and 47 % relative abundance, respectively), whereas the control formula lagged behind. Crucially, the relative abundance of Bifidobacterium in the 2′-FL arm was statistically indistinguishable from breast-fed infants, demonstrating a robust bifidogenic effect that standard prebiotics alone did not achieve.
Beyond genus-level shifts, the 2′-FL formula proved safe and well tolerated, supporting normal growth trajectories while shaping a gut community that, although more diverse than breast milk, leaned toward breast-like functional hallmarks. These findings suggest that adding specific HMOs can help bridge the microbial gap between human milk and formula, potentially delivering downstream benefits for immune training and metabolic programming. As HMO-fortified formulas proliferate, studies like this provide a mechanistic microbiome blueprint for designing next-generation infant nutrition that more faithfully echoes nature’s original recipe.
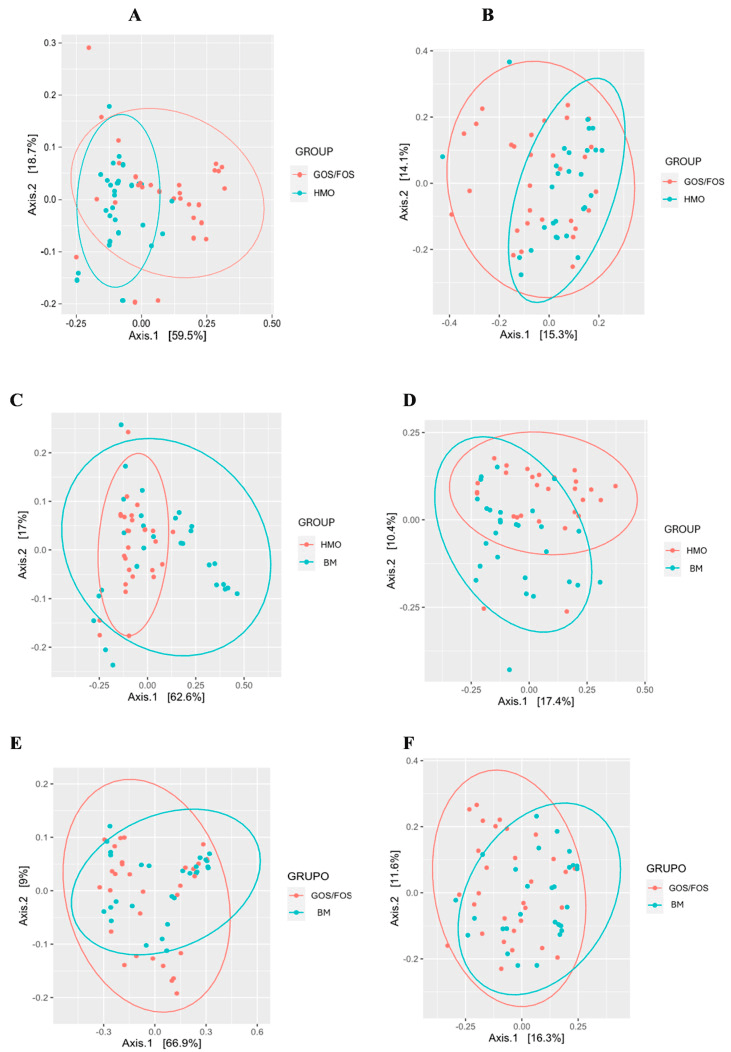
Beta-diversity: pairwise comparison between groups after intervention. (*) = statistical significance. Panel (A) UniFrac distance weighted at the end according to HMO x GOS/FOS groups p = 0.002 *. Panel (B) Final unweighted UniFrac distance for HMO vs. GOS/FOS groups p = 0.013 *. Panel (C) Final weighted UniFrac distance for HMO vs. BM groups p = 0.009 *. Panel (D) Final unweighted UniFrac distance for HMO vs. BM groups p = 0.001 *. Panel (E) Final weighted UniFrac distance according to the GOS/FOS x BM groups p = 0.185 *. Panel (F) Final unweighted UniFrac distance for GOS/FOS vs. BM groups p = 0.002 *. HMO, experimental group; GOS/FOS, control group; BM, Breast milk-fed infants.
Gut Diversity Tips the Balance in Triple-Negative Breast Cancer Immunotherapy
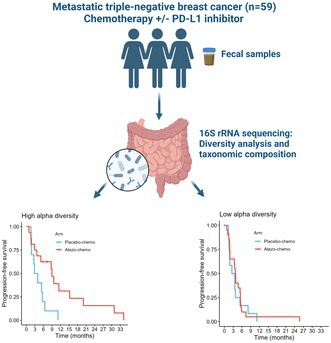
Triple-negative breast cancer (TNBC) has joined the growing list of malignancies where the gut microbiome appears to tip the scales of immune therapy. In the ALICE phase II trial, Norwegian investigators paired atezolizumab with chemotherapy and then sequenced stool collected before treatment and after eight weeks to see whether microbial patterns tracked with clinical outcome. Their focus on microbiome diversity is timely: diverse gut ecosystems are thought to supply a broader repertoire of immunostimulatory metabolites and antigens that can prime systemic antitumor immunity.
Baseline results were striking. Patients whose guts harboured high phylogenetic diversity (Faith’s PD above the cohort median) enjoyed markedly longer progression-free survival, an effect that was specific to the atezolizumab + chemo arm (hazard ratio 0.34, P = 0.018) and absent in chemo alone (HR 0.83, P = 0.62). Diversity fell in both arms during treatment—likely collateral damage from cytotoxic drugs—yet the prognostic signal of the starting microbiome remained. These data position alpha diversity not merely as a bystander metric but as a potential companion biomarker to select candidates most likely to benefit from chemo-immunotherapy.
Taxonomic clues added nuance: Bifidobacterium, often celebrated as a probiotic hero, was paradoxically enriched in atezolizumab-treated patients who failed to respond, reminding us that context matters and that single-genus “goods” may not translate across disease settings. Together, the study argues for preserving or even augmenting gut diversity—perhaps through diet, prebiotics, or rational microbial consortia—before immune-checkpoint blockade begins. As microbiome-driven precision oncology advances, the gut may become as testable and tunable as the tumour itself.
Spores vs. Spoilage: A Probiotic Strategy to Preserve Richness in C. difficile Treatment
Vancomycin clears Clostridium difficile, but it also mows down the commensal flora that normally keep this pathogen in check. In a multicenter Korean pilot study, clinicians tried a different tack: they paired the antibiotic with a five-day course of the spore-forming probiotic Bacillus licheniformis and then tracked how the gut community fared. Thirty-five adults with mild-to-moderate CDI provided stool before treatment and again on day 5. While diversity (Chao1, observed features) fell sharply in the placebo arm, it held steady in those who received the probiotic—essentially insulating microbial richness against vancomycin’s collateral damage.
The protective signal emerged fast: by the first sampling point, alpha-diversity in the probiotic group was statistically unchanged (p ≈ 0.7), whereas the placebo group lost significant richness (p = 0.011). Beta-diversity shifts were comparable between groups, suggesting that B. licheniformis functions less as a niche-replacing colonizer and more as a resilience aid, helping resident taxa rebound after antibiotic shock. Discussion notes highlight why this matters: deeper dysbiosis predicts both CDI recurrence and post-infectious IBS, so holding the line on diversity—even for a few crucial days—could translate into fewer relapses and better long-term gut health.
Mechanistically, spore-based bacilli weather gastric acid and germinate downstream, where they may secrete antimicrobials, compete for nutrients, or simply occupy ecological space until obligate anaerobes recover. Unlike fecal microbiota transplantation—which carries logistical and safety hurdles—this single-strain approach is cheap, shelf-stable, and easy to deploy alongside standard care. The authors caution, however, that their sample was small and follow-up brief; larger trials will have to confirm whether early preservation of diversity actually lowers CDI recurrence rates. Still, the data offer a microbiome-centric proof-of-concept: sometimes protecting the gut’s species richness is as important as eradicating the pathogen that upset it in the first place.
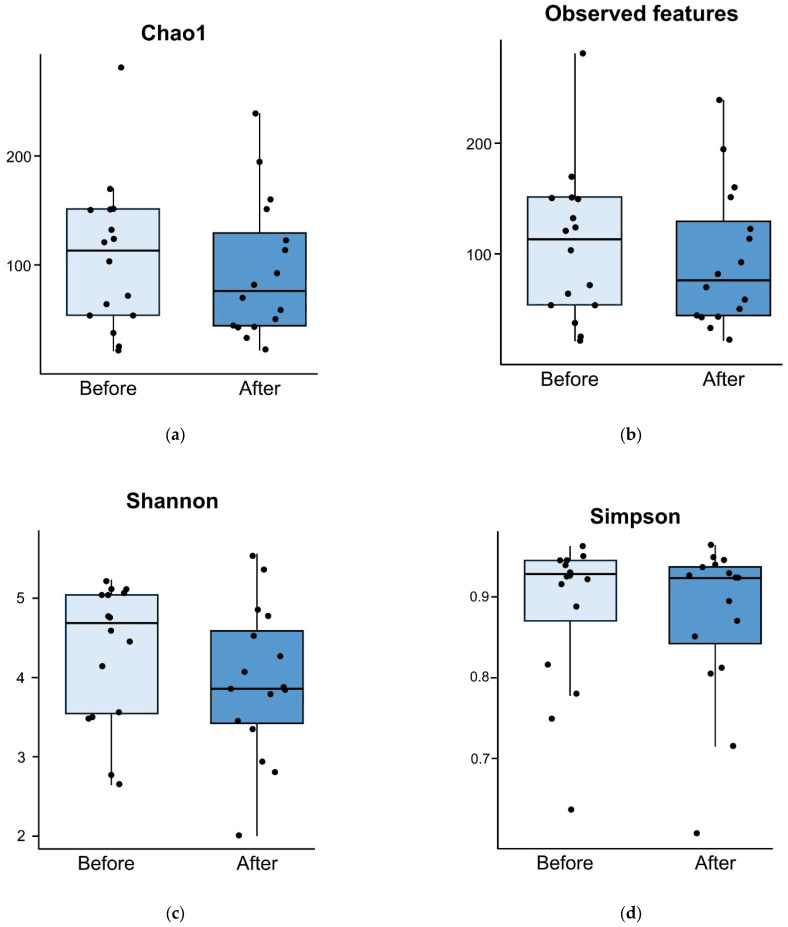
Comparison of alpha diversity for probiotic group between before and five days after treatment. Alpha diversities measured using Chao1 index (p = 0.665) (a), observed features (p = 0.692) (b), Shannon diversity index (p = 0.287) (c), and Simpson diversity index (p = 0.806) (d) are comparable before and after treatment.
