Holobiome: Research I Articles I Updates I News I Series I Feed
Holobiome is a blog series that offers an AI-assisted summary of the latest research articles on human microbiome.
Vertical Transmission Reimagined: How Maternal Probiotics Strengthen Infant Gut Networks
Pregnancy and early life mark a crucial window for shaping the infant microbiome, a microscopic inheritance that profoundly influences immunity and metabolism. In a recent clinical trial, researchers explored whether a probiotic supplement given to mothers during late pregnancy and breastfeeding could tip the scales toward a healthier microbial start for their babies. The intervention combined two well-studied species, Lacticaseibacillus rhamnosus Rosell®-11 and Bifidobacterium bifidum HA-132, taken daily at roughly five billion CFU.
While the probiotic didn’t drastically alter the mothers’ vaginal microbiome, its influence emerged in subtler and more biologically intriguing ways. Infants born to supplemented mothers, especially those delivered by Caesarean section, showed gut communities richer in beneficial microbes, an encouraging sign, since C-section delivery often limits natural microbial transfer from mother to child. The study’s analyses revealed that probiotic intake improved the “vertical transmission” of certain bacteria through breast milk, effectively helping newborns acquire the microbial allies they might otherwise miss.
The researchers also used advanced network modeling to visualize how these microbes connected and co-occurred within the infant gut. In babies whose mothers took probiotics, the microbial ecosystem appeared more cohesive, with Bifidobacterium at the center of a tightly linked community suggesting a more stable and resilient gut network. Such patterns are not just academic; stronger microbial networks in infancy have been linked to lower infection risk and better immune training.
Taken together, these findings highlight the quiet power of maternal probiotics: rather than overhauling the microbiome, they fine-tune the transfer and assembly of key bacterial players during a pivotal developmental window. By subtly reinforcing microbial hand-offs from mother to child, this approach could help lay a healthier microbial foundation one that supports early immune resilience and long-term well-being.
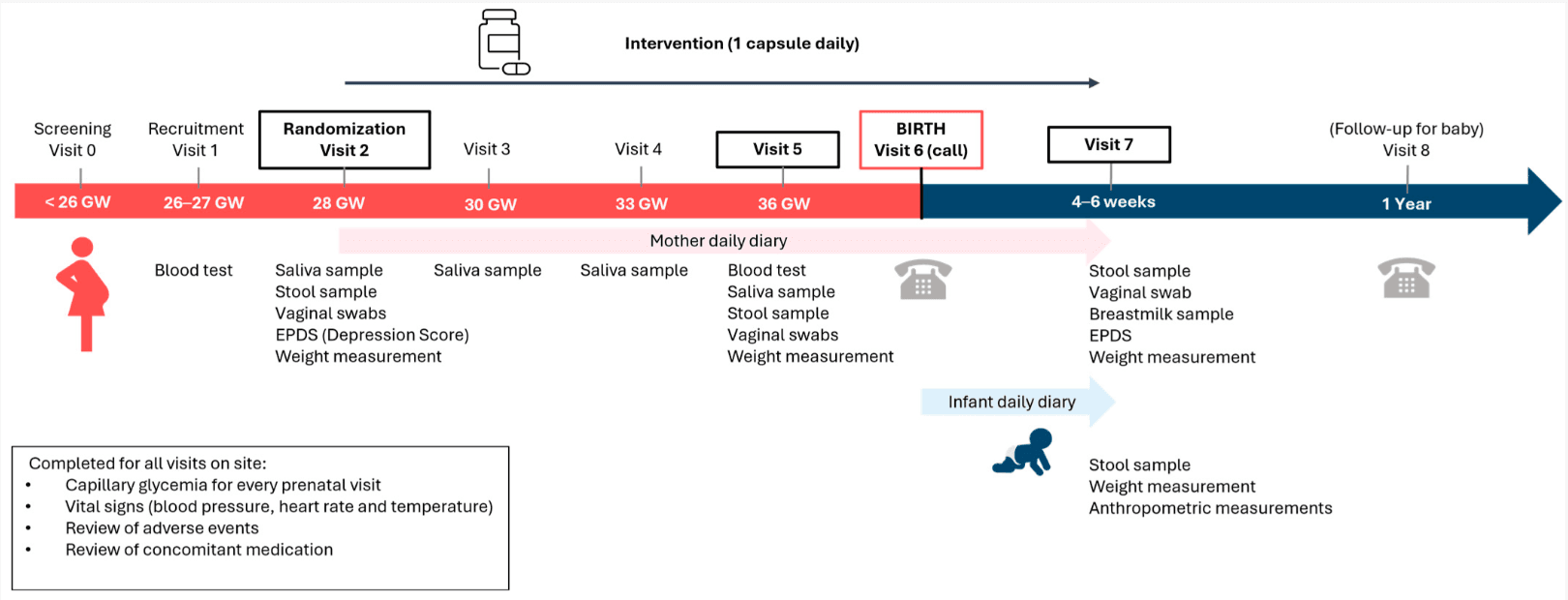
Study design. The study was a randomized, double-blind, placebo-controlled, parallel-arm trial spanning an 18-week period. The sample size for this study was 201 participants enrolled, with 180 participants randomized in a 1:1 ratio in each of the 2 arms. This study consisted of 8 visits, with prescreening, recruitment, and randomization visits, 5 interim visits, and a final end-of-study visit. To evaluate the primary and secondary outcomes, study assessments were conducted in 8 visits.
Balancing Eradication and Ecology: How Amoxicillin Dose Shapes the Gut Resistome After H. pylori Therapy
The study by Hu et al. delves into a fascinating junction of microbiome science, antibiotic therapy, and the hidden costs of microbial disruption. The authors compared two regimens of Helicobacter pylori eradication a dual therapy combining vonoprazan with either a lower amoxicillin dose (2 g/day) or a higher dose (3 g/day) and tracked not only how well the bacteria were cleared, but how the gut microbiome and its antibiotic-resistance genes responded.
Before treatment, participants had characteristic gut microbial communities. After the 14-day therapy, both arms showed a marked drop in microbial diversity in stool, reflecting the collateral damage of antibiotics. However, by weeks 8–10, the diversity largely rebounded to baseline levels in both groups, a sign of resilience in the gut ecosystem. What distinguished the two arms was the dynamic of antibiotic resistance genes, especially those linked to beta-lactams: the higher dose group retained an elevated abundance of resistance genes even at the 8–10 week follow-up, while the lower dose group’s resistome more fully reverted toward its original state.
From a microbiome lens, the trial is a reminder that eradication therapies, even when targeted, ripple through the microbial community. It’s not just about hitting H. pylori the antibiotics also perturb other bacterial taxa, suppressing some and allowing temporary blooms (or losses) in others. The transient loss in richness suggests that some rarer or sensitive species are knocked back during therapy. Then, as recovery proceeds, competitive dynamics and niche reoccupation determine which microbes re-emerge. In the lower-dose arm, the resistome’s recovery suggests that “collateral awakenings” of resistance genes can be better contained with gentler antibiotic pressure.
Ultimately, the authors show that a lower amoxicillin dose is non-inferior in clearing H. pylori, yet causes less lasting disturbance to the gut resistome. For those of us fascinated by microbiome ecology, it's a useful case study: microbial communities are buffeted by clinical interventions, but the degree of disturbance and the speed (or completeness) of recovery depend on how much pressure we exert. Thoughtful dosing choices, then, can help us strike a balance between therapeutic goals and microbial stewardship.
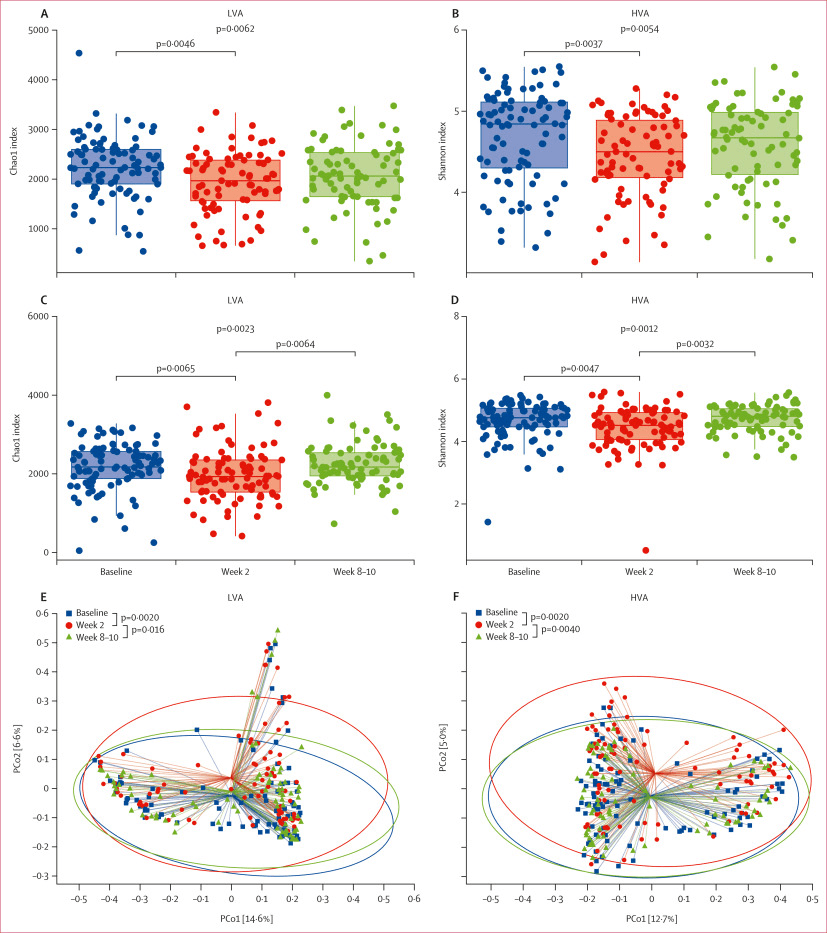
Alpha diversity estimated by Chao1 (A, C) and Shannon (B, D) indices of the gut microbiota at baseline, week 2, and week 8–10 in the LVA therapy (A, B) and HVA therapy (C, D) groups. Beta diversity estimated by principal coordinates analysis of the gut microbiota at baseline, week 2, and week 8–10 in the LVA therapy (E) and HVA therapy (F) groups. HVA=high-dose amoxicillin and vonoprazan. LVA=low-dose amoxicillin and vonoprazan. PCo=principal coordinate.
Doxycycline Prophylaxis and the Gut Resistome : What Happens Beneath the Surface
The trial explored what happens to your gut microbial community when you use doxycycline post-exposure prophylaxis (doxy-PEP) a strategy that some people take to reduce their risk of bacterial STIs. The researchers enrolled adult participants (men who have sex with men and transgender women) and monitored fecal samples before and after doxycycline use, seeking to understand how the gut microbiome and its reservoir of antibiotic resistance genes (the “resistome”) respond.
Across participants, doxycycline use led to measurable shifts: species richness and microbial diversity dropped shortly after exposure reflecting the well-known “collateral damage” antibiotics inflict on the gut ecosystem. Some taxa declined more than others, especially sensitive or low-abundance microbes, creating room for subtle reorganizations in the community. However, the changes were not uniform: individuals diverged in how fast and how fully their microbiota rebounded. Importantly, while many taxa recovered at later time points, the resistome showed lasting perturbations. Certain antibiotic resistance gene families especially those linked to tetracycline and broader classes were enriched after doxycycline use and lingered longer than the taxonomic disruptions.
From a microbial ecology vantage, this trial illustrates how introducing antibiotic pressure even intermittently can tilt the balance between microbial resilience and resistance. The microbiota may recover structural measures like diversity, yet the hidden baggage of antimicrobial resistance may persist, altering the “memory” of the gut ecosystem. That means even when your gut seems “back to normal,” its genetic arsenal might have shifted. What the authors emphasize is that doxy-PEP’s benefits must be weighed against these subtle, longer-lasting effects on the gut microbiome and resistome. In short, the trial underscores a delicate tradeoff: using antibiotics as preventive tools carries ecological costs in the microbial world, and each dose adds a whisper to the evolving story of your gut’s microbial legacy.
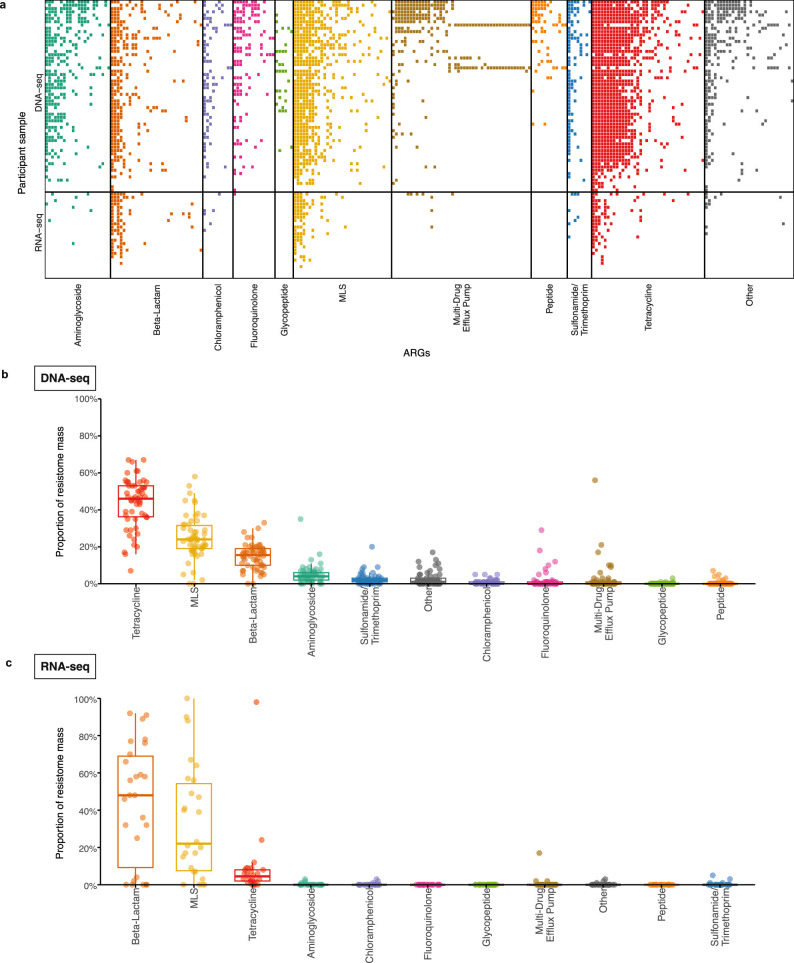
a) Heatmap of the antimicrobial resistance genes (ARGs) detected in DNA-seq samples (n = 58) and RNA-seq samples (n = 26) at enrollment by ARG class. Proportion of the resistome mass by ARG class on enrollment in (b) DNA-seq samples (n = 58) and (c) RNA-seq samples (n = 26). Boxplot elements include a center line (median), box limits (upper and lower quartiles), whiskers (1.5x interquartile range). Abbreviation: ARG, antimicrobial resistance gene; MLS, macrolide-lincosamide-streptogramin.
When More Isn’t Better: The Limits of Microbiome-Directed Antibiotic Therapy in Cystic Fibrosis
In cystic fibrosis, the lungs host a complex community of microbes that both sustain and sabotage respiratory health. The CFMATTERS clinical trial set out to test whether precision-guided antibiotic therapy based on each patient’s sputum microbiome could outperform standard treatment during pulmonary exacerbations. Across nine international centers, 223 adults with chronic Pseudomonas aeruginosa infection were enrolled and randomized: one group received standard dual intravenous antibiotics (ceftazidime + tobramycin), while the other received the same regimen plus a third antibiotic tailored to the second, third, and fourth most abundant genera detected through next-generation sequencing of sputum samples.
The microbial logic behind this approach was compelling. Cystic fibrosis airways rarely harbor a single culprit; instead, polymicrobial biofilms dominated by Pseudomonas coexist with genera like Staphylococcus, Streptococcus, Burkholderia, Achromobacter, Prevotella, and Veillonella. Many of these secondary or anaerobic taxa can fuel inflammation or modulate antibiotic response. By using DNA sequencing to identify these hidden contributors, researchers hoped that an additional, microbiome-directed antibiotic might better clear infection and improve lung function.
Yet biology proved more nuanced. Lung function (measured by ppFEV1) improved similarly in both groups at day 14, and no significant differences emerged at later checkpoints or in time-to-next exacerbation. Intriguingly, those receiving microbiome-guided therapy experienced slightly more exacerbations and intravenous antibiotic days over the following year. Microbial analyses revealed that while Pseudomonas abundance decreased with treatment as expected the overall airway community composition largely rebounded to its pre-treatment state within weeks. In other words, the microbiome seemed resilient to both strategies, re-assembling into its original ecological balance despite additional antimicrobial pressure.
From a microbiome perspective, the trial delivers an important reality check. The cystic fibrosis lung microbiome behaves like a resilient ecosystem rather than a collection of simple targets. Simply adding antibiotics based on genus abundance does not guarantee better outcomes and may even disrupt delicate microbial equilibria. The findings highlight the need for next-generation approaches that go beyond cataloging taxa to understanding microbial function, virulence, and inter-species interactions. The message is clear: in the microbial landscape of cystic fibrosis, precision therapy will require more than just deeper sequencing; it will demand ecological insight.
