Holobiome: Research I Articles I Updates I News I Series I Feed
Holobiome is a blog series that offers an AI-assisted summary of the latest research articles on human microbiome.
Microbes on their own time: How the gut responds to eating windows
In this study, researchers revisited a 12-week randomized controlled trial of time-restricted eating (TRE) in adults with overweight or obesity, drilling down into stool samples to explore how the gut microbial community might respond to limiting food intake to an 8-hour window each day. Participants in the TRE arm narrowed their eating window from around 15 hours to just over 9 hours and, as expected, lost weight and visceral fat—changes that raised the question: did the microbes in the gut shift too?
Surprisingly, the gut microbiome appeared remarkably stable. Measures of alpha diversity (which capture species richness, evenness, and diversity) and beta diversity (which compare overall community composition between timepoints or individuals) showed no significant shifts in the TRE group compared to controls, even after adjusting for weight or fat loss. When zooming in on the handful of participants with both pre- and post-intervention stool samples, the authors also saw no consistent trends in the abundance of specific species or genera, suggesting a high level of individual variation and perhaps no clear “microbial signature” of TRE in this small sample.
The researchers caution that the small sample size, limited paired sampling, and short timeframe likely limited their ability to detect more subtle microbial shifts. They also note that functional changes—such as in microbial metabolism or circadian rhythms of the microbiome—could be happening without large taxonomic turnover, and that timing of stool collection might have missed dynamic daily fluctuations. The take-home: in this pilot TRE trial, the gut microbiome showed surprising resilience in taxonomic structure over 12 weeks, but more in-depth, time-sensitive sampling and functional analyses are needed to know whether TRE might subtly rewire microbiome behavior even if the lineup of species stays much the same.
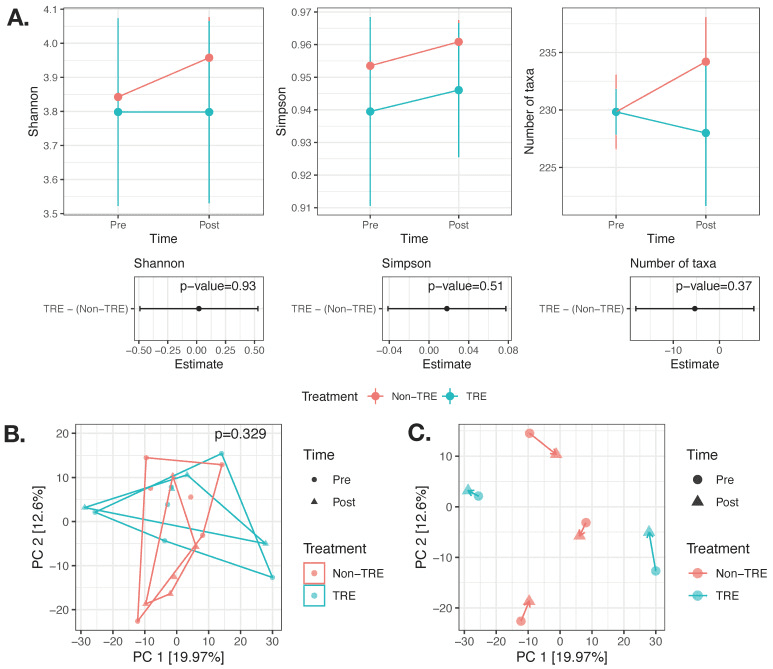
No significant change in alpha or beta diversity after 12 weeks of TRE. (A) The effect of the TRE intervention on alpha diversity (Shannon, Simpson, and the number of taxa) was modeled using linear mixed models adjusted for weight change and visceral fat change and accounting for within-subject repeated measures. n = 21 samples from 16 participants (8 TRE and 8 Non-TRE). (B) Beta diversity modeled using PERMANOVA and adjusted for weight change and visceral fat change and accounting for within-subject repeated measures. n = 21 samples from 16 participants (8 TRE and 8 Non-TRE). (C) Visualization of the paired subset of samples (n = 10 samples from 5 participants) did not reveal clear trends within study arms with respect to directionality of changes over the first principal coordinates. TRE: time-restricted eating; Non-TRE: time-unrestricted eating, PC: principal coordinate.
The Microbiome’s Role in Surgical Recovery: A Tale of Lactic Acid and Friendly Bacteria
In this large exploratory study, the authors investigated whether features of the gut microbiome and fecal organic acid profiles, measured before surgery, could help predict which patients will go on to develop postoperative infections after major gastrointestinal cancer operations. They analyzed stool samples from 381 patients via 16S rRNA gene sequencing and fecal organic acid quantification, aiming to link preoperative microbial and metabolic markers to later infectious complications.
One striking finding was that elevated fecal lactic acid levels before surgery stood out as a strong independent risk factor for infectious complications—including pneumonia, wound infections, abscesses, cholangitis and sepsis—suggesting that higher lactic acid reflects a disrupted gut environment that may predispose to infection. In contrast, two microbial species—Akkermansia muciniphila and Lactococcus lactis—were more abundant in patients who did not go on to have postoperative infections. These species appear to correlate with better gut barrier function and immune resilience. A. muciniphila in particular is thought to strengthen the gut’s mucosal lining and modulate immune responses, potentially reducing bacterial translocation during the stress of surgery.
Although the study didn’t find a direct link between lactic acid levels and the abundances of A. muciniphila or L. lactis, the authors propose that high lactic acid may instead reflect broader dysbiosis—potentially due to excessive production by lactic acid–producing bacteria or reduced consumption by lactic acid-utilizing microbes—combined with impaired gut barrier and metabolic conditions in cancer patients. Overall, the results suggest that measuring fecal lactic acid and microbial composition preoperatively could become a useful strategy for identifying patients at higher risk of post-surgical infections, and that boosting beneficial taxa like A. muciniphila or L. lactis might someday form part of microbiome-targeted preventive strategies for surgical patients.
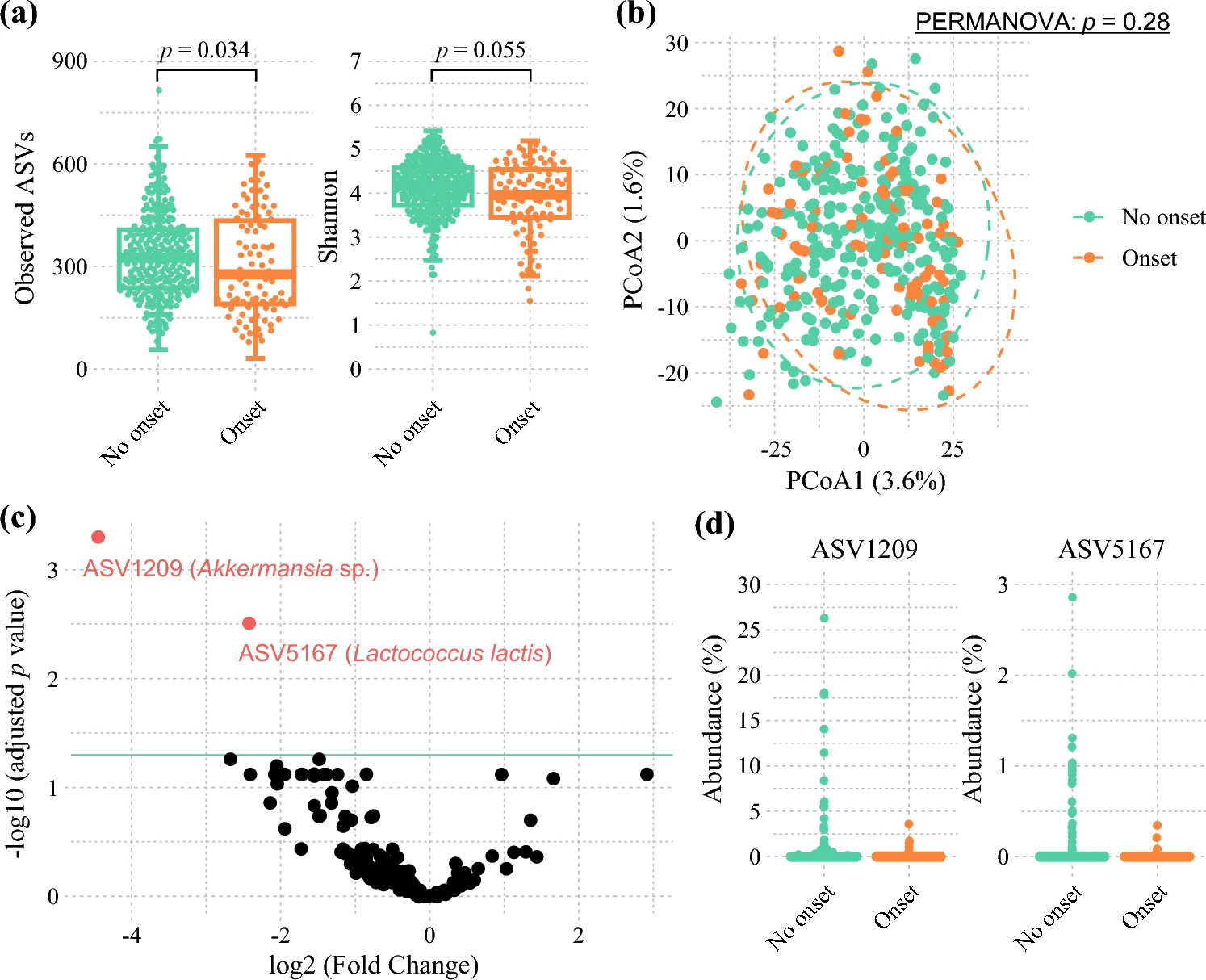
Comparison of the preoperative gut microbiota between patients who had postoperative infectious complications and those who did not. a Alpha diversity and b beta diversity based on the Aitchison distance compared between patients who had postoperative infectious complications and those who did not. For alpha diversity, p values were estimated using the Wilcoxon rank-sum test. For beta diversity, p values were estimated using the pairwise permutational analysis of variance (PERMANOVA) test. c Volcano plot of differentially abundant taxa between patients who had postoperative infectious complications and those who did not, assessed using DESeq2. Black points represent amplicon sequence variants (ASVs) with no significant difference in abundance, and red points represent ASVs with a significant difference in abundance (p < 0.05). d The abundance of significantly different taxa using DESeq2 compared between patients who had postoperative infectious complications and those who did not.
Colon Tissue Microbiome: Silent Players in Adenoma Recurrence
This study takes a close look at the microbial communities residing in colon tissue—rather than stool—to see how their composition might be linked to the recurrence of colorectal adenomas (precursor lesions to cancer). By pairing tissue-based microbiome profiling with measurements of circulating bile acids, the authors aimed to tease out how microbial shifts and metabolite changes might influence adenoma outcomes.
One of the key observations was that differences in the relative abundance of major bacterial phyla—especially Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes—helped distinguish groups of patients with different adenoma recurrence risks. Patients whose colon tissues showed higher Proteobacteria and Bacteroidetes and a lower Firmicutes: Bacteroidetes ratio were more likely to experience recurrence, hinting at a link between a dysbiotic tissue microbiome and adenoma risk. In contrast, tissue microbiomes with greater representation of Actinobacteria and Firmicutes seemed more characteristic of patients with better outcomes.
The study proposes that microbes living in or near tumor tissue might influence adenoma recurrence through metabolite production and interactions with bile acids—possibly affecting inflammation, epithelial cell proliferation, and local immune responses. Although the authors acknowledge limitations (such as lacking healthy control tissue and the challenge of discerning cause versus effect), the findings suggest that profiling the colon tissue microbiome could one day help stratify recurrence risk. In short, not all microbes are merely bystanders: those living in the colon wall may actively shape whether adenomas come back, and they may serve as informative biomarkers or even therapeutic targets in colorectal cancer prevention.
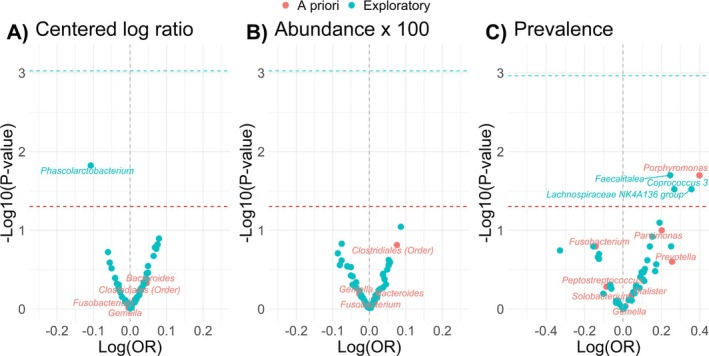
Volcano plot of the associations of bacteria with adenoma among N = 165 adenoma cases and N = 311 controls in the Colorectal Neoplasia Screening with Colonoscopy in Average‐Risk Women Regional Navy/Army Medical Centers study, 2000–2002; bacteria abundances were characterized using the (A) standardized centered log ratio transformation, (B) standardized relative abundance x 100, or (C) presence/absence of the bacterium; conditional logistic regression models adjusted for age, year of colonoscopy, regular nonsteroidal anti‐inflammatory drug and/or aspirin use (yes/no), current alcohol use (yes/no), family history of polyps (yes/no), education (high school education or less; or some college or more), hormone replacement therapy use (yes/no), daily fiber intake (g/day), body mass index (kg/m2), total daily energy intake (kcal/day), physical activity level (no moderate or vigorous activity, unknown, missing; moderate activity, or vigorous activity), race (Black, White, other), smoking status (current/former smokers or never smokers), red and processed meat intake (g/day), and study center (Walter Reed, San Diego, Portsmouth, or Bethesda); all bacteria are at the genus level except for order Clostridiales; we used Bonferroni correction to account for multiple testing the number of statistical comparisons (i.e., alpha level = 0.05/53 comparisons for abundance and 0.05/46 for prevalence) for all exploratory analyses (blue dotted line) and an alpha level of p = 0.05 for the a priori analysis (red dotted line).
The Rapidly Changing Gut: How Infants’ Microbes Evolve Day by Day
This study tracked how the gut microbiome of healthy infants develops over the first year of life—with an especially detailed focus on how sampling frequency influences what we think we know. One infant (A) provided near-daily stool samples, while twelve others (B–M) were sampled only weekly. The researchers used 16S rRNA gene sequencing to profile the bacterial communities, capturing the dynamic ebb and flow of early colonizers, re-appearing microbes, and later-arriving taxa.
What emerged was a vivid picture of microbial plasticity: frequent sampling revealed rapid fluctuations in alpha diversity (within-sample richness and evenness) that weekly sampling completely missed. Within the first 23 weeks, samples taken just days apart could differ by more than one Shannon diversity unit—suggesting that early gut communities are extremely dynamic. Interestingly, as infants aged, alpha diversity variation calmed down, but beta diversity (the differences between samples over time) stayed surprisingly high—a sign that while overall richness stabilizes, the composition of gut communities continues to shift.
The study also tied major life events—such as the introduction of solid foods, probiotic supplementation, or vaccinations—to gradual yet meaningful changes in microbial composition. These shifts looked different from one infant to another, underlining the strong individual signature in gut microbiome development. Weekly sampling, the authors warn, can under-capture critical transitions in colonization trajectories, potentially masking how early microbial exposures shape long-term gut ecosystem trajectories.
Overall, the work underscores that to truly understand how infant gut microbiomes assemble—and how early life exposures shape lifelong microbial ecosystems—sampling frequency matters a lot. A once-a-week snapshot might miss a host of microbial dramas playing out in the days in between.
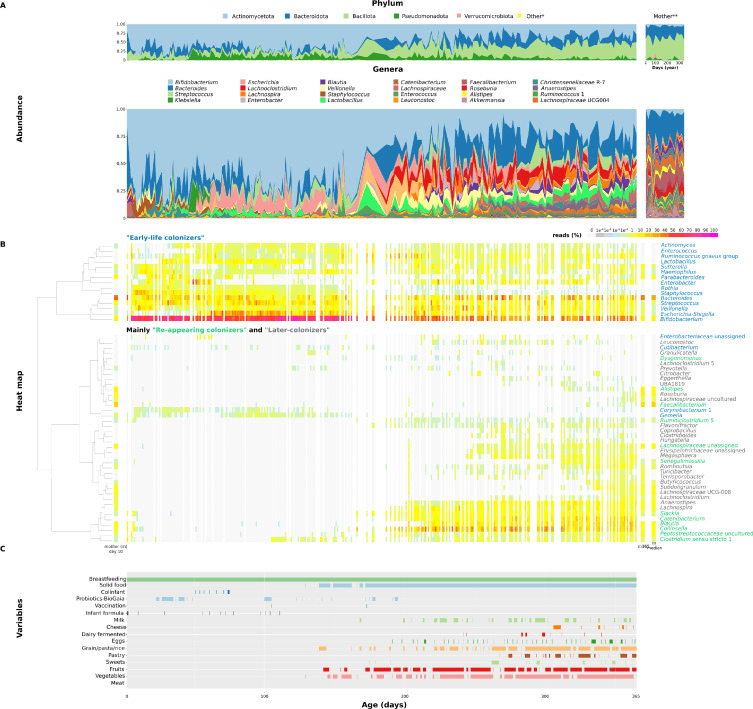
Composition of infant A's and mother's fecal microbiome during the first year of life. A. Dynamics of the fecal microbiome at the phylum or genus level. *Bacteria with a maximal incidence below 1 %. **Mother's stool samples collected throughout the year. B. Heatmap dendrogram showing the co-occurrence of 55 bacterial genera (genera present in at least 2 % of samples with a minimum abundance of 10 reads) in time. Clustering was made using Ward's method on Euclidean distances of CLR-normalized bacterial abundance data. White columns indicate missing samples. C. Main factors/variables/events monitored during the year.
From Nitrate to Nitric Oxide: Microbes That Power Performance
This study turned a spotlight on the oral microbiome—especially the tongue dorsum—to see how elite athletes’ mouths differ from those of inactive individuals. Using long-read 16S rRNA sequencing, the researchers found that athletes had higher abundances of Rothia mucilaginosa and unclassified Gemella species on the tongue, both of which are known nitrate-reducing microbes capable of converting salivary nitrate (NO₃⁻) to nitrite (NO₂⁻)—a key step in the entero-salivary nitric oxide (NO) production pathway.
These microbial shifts were mirrored in biochemical measurements: athletes showed elevated salivary nitrate and nitrite levels, suggesting their oral microbiome might be more actively contributing to NO production, which has implications for vascular, oral, and systemic health. Interestingly, the supragingival plaque microbiome didn’t show major differences between athletes and controls, highlighting that the tongue dorsum might host a more responsive microbial niche when it comes to exercise-related shifts.
The study paints a compelling portrait of how regular aerobic training might subtly nurture an oral microbial ecosystem—especially taxa involved in nitrate-nitrite conversion—that supports NO bioavailability and possibly oral and cardiovascular resilience. Whether these shifts translate into long-term health benefits or reduced dental disease risk remains to be seen, but the findings suggest the oral microbiome may serve as a hidden marker of fitness or even a target for probiotics aimed at enhancing NO-related pathways.
