Holobiome: Research I Articles I Updates I News I Series I Feed
Holobiome is a blog series that offers an AI-assisted summary of the latest research articles on human microbiome.

Beneath the Gums: How Microbial Communities Shape Periodontal Disease Progression
Periodontal health depends on a finely balanced subgingival microbiome, and this study shows just how dramatically that balance shifts as disease develops and accelerates. Using deep metagenomic sequencing, the authors compared healthy individuals with two forms of severe periodontitis that differ in how quickly they progress. Even without obvious clinical differences, the microbial ecosystem beneath the gumline told a much richer story one defined by dysbiosis, microbial cooperation, and functional reprogramming.
In health, the subgingival niche was dominated by bacteria typically associated with stability and biofilm organization, such as Corynebacterium matruchotii, Actinomyces naeslundii, and Cardiobacterium hominis. These microbes were negatively correlated with inflammation and tissue damage, suggesting a protective or homeostatic role. In contrast, periodontitis samples showed an expansion of classic periodontal pathogens, including Porphyromonas gingivalis, Treponema denticola, Tannerella forsythia, and Prevotella intermedia. This shift reflects a move away from a structured, health-associated community toward a more aggressive, inflammation-linked microbiome.
A particularly striking finding was that rapidly progressing periodontitis harbored subtle but meaningful microbial differences. Two Capnocytophaga species Capnocytophaga granulosa and Capnocytophaga sp. CM59 were enriched in the fast-progressing group and helped distinguish it from more slowly advancing disease. Network analyses revealed that these severe cases were marked by denser, more positively connected microbial networks, suggesting that pathogens may cooperate rather than compete as disease worsens.
Functionally, the diseased microbiome appeared primed for invasion. Genes linked to bacterial motility, chemotaxis, methane metabolism, and sulfur metabolism were enriched in periodontitis, especially in rapidly progressing cases. In contrast, health-associated communities emphasized core metabolic maintenance. Together, these findings reinforce the idea that periodontitis is not driven by single pathogens, but by coordinated microbial communities whose structure and function evolve alongside disease severity opening the door to microbiome-informed diagnostics and more personalized periodontal care.
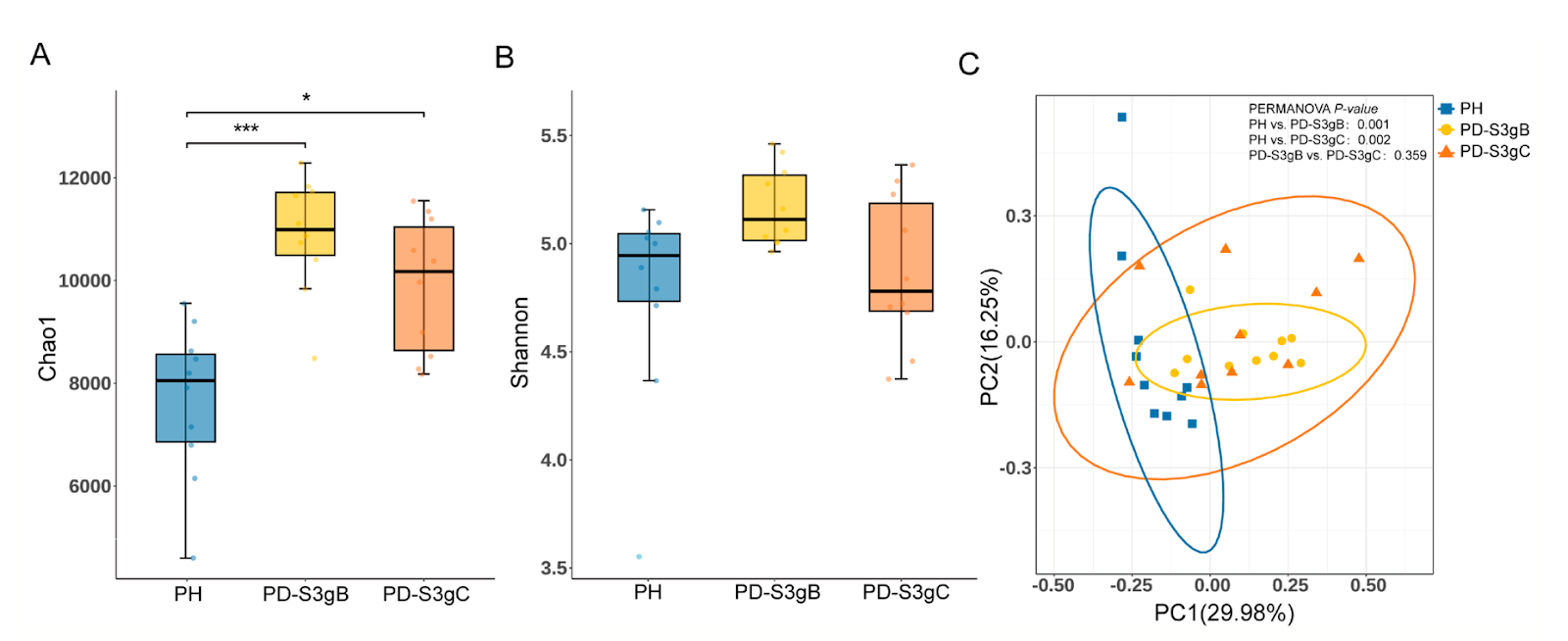
FIGURE 1 | Microbial diversity of the subgingival community at the species level in three periodontal conditions. A. and B. Chao1 index (A) and Shannon index (B) of the alpha diversity. Data were analyzed using the Kruskal–Wallis H test and Dunn's multiple comparisons test with BH adjustment (*p < 0.05, **p < 0.01, ***p < 0.001). (C) Principal coordinates analysis (PCoA) plot (based on Bray- Curtis distance) and PERMANOVA result of the microbiome structure. PD- S3gB, Stage III Grade B periodontitis; and PD- S3gC, Stage III Grade C periodontitis; PH, periodontal health.
How Gut Microbes Shape Chemotherapy Outcomes in Brain Lymphoma
This study offers a compelling look at how the gut microbiome may shape treatment outcomes in a rare but aggressive brain cancer, primary central nervous system lymphoma (PCNSL). While therapy for PCNSL relies heavily on high-dose methotrexate–based chemotherapy, patients respond very differently, and this work explores whether intestinal microbes help explain why. By combining metagenomic sequencing of stool samples with immune profiling and plasma metabolomics, the researchers traced a biological conversation between gut bacteria, circulating metabolites, and immune activity in the central nervous system.
At the center of this story is a single bacterial species, Parabacteroides distasonis. Patients naturally clustered into two gut microbiome community types, and those enriched in P. distasonis showed significantly longer progression-free and overall survival. This association was not explained by general microbial diversity, diet proxies like body mass index, or chemotherapy timing. Instead, P. distasonis appeared to act as a functional keystone, shaping metabolic outputs and immune signaling rather than simply altering which microbes were present.
One important clue came from metabolomics. Higher levels of P. distasonis were linked to increased plasma concentrations of a metabolite signature centered on betaine–valine. This metabolic pattern independently predicted better clinical outcomes in a larger validation cohort, suggesting that microbial metabolism, not just microbial presence, matters. Metagenomic pathway analysis further showed enrichment of amino acid biosynthesis pathways connected to P. distasonis, reinforcing the idea that this bacterium actively contributes to the host’s metabolic environment.
The immune system provided the final link. Patients with high P. distasonis levels had greater infiltration of CD8⁺ T cells into the cerebrospinal fluid, pointing to a gut–brain–immune axis in PCNSL. Together, these findings suggest that specific gut microbes may prime systemic immunity and improve chemotherapy effectiveness. While still early, the work opens the door to microbiome-informed patient stratification and raises the possibility that future supportive strategies such as targeted prebiotics or microbiome-preserving interventions could help tilt outcomes in favor of patients facing this challenging disease.
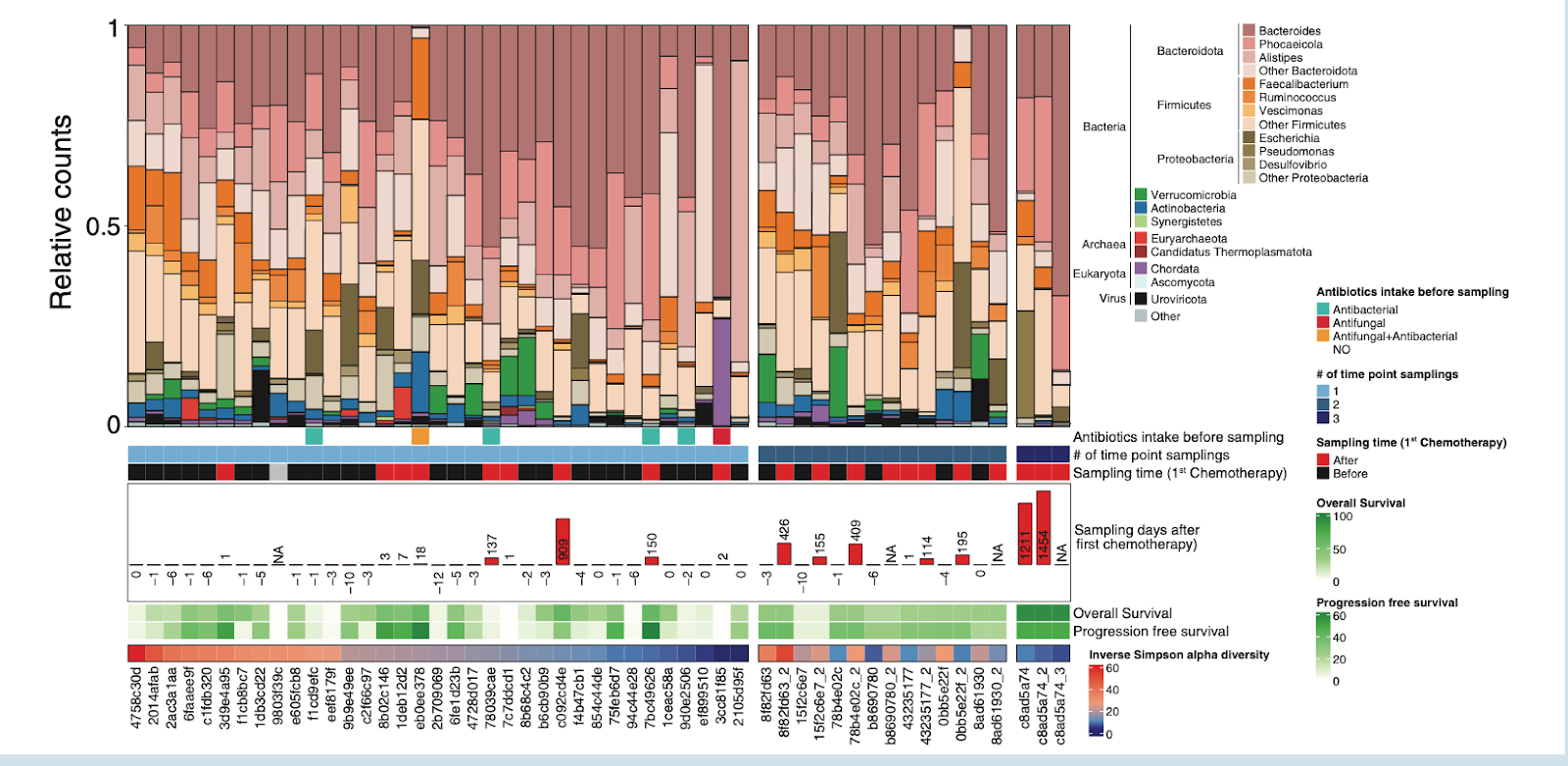
Figure 2. position of fecal samples (n only 1 sampling point (n Distribution of microbiome composition in primary central nervous system lymphoma patients. Barplot showing the phylogenetic com = 52). Samples are ordered by decreasing inverse Simpson alpha diversity values and by a group of patients having = 35, left), 2 sampling points (n = 7), or 3 sampling points (n= 1).
Microbial Clues in Acute Pancreatitis: What the Gut Reveals Early On
When people are hospitalized with acute pancreatitis a sudden, painful inflammation of the pancreas their gut microbiome doesn’t just sit quietly in the background. This study explored whether the composition of microbes at the time of hospital admission could be linked with how severe the disease becomes and whether complications develop later. By sequencing the bacterial DNA from rectal and saliva samples of hundreds of patients, the researchers captured a snapshot of each person’s microbial community as they first entered care. What they found paints a picture of how gut ecology might influence systemic disease processes.
One clear pattern was that patients with more severe, necrotizing pancreatitis tended to have lower microbial diversity in their gut. In general, diversity the number and evenness of bacterial species is considered a hallmark of a resilient, health-promoting microbiome. Lower diversity has been linked in many contexts to dysbiosis, a state where protective species are lost and opportunistic or pathogenic taxa can rise.
Indeed, several specific bacterial groups stood out. Three taxa Finegoldia, Anaeroglobus, and a group within the Lachnospiraceae family were robustly associated with more severe disease features. Although their exact roles aren’t fully understood, the recurrence of Lachnospiraceae is intriguing because many of its members are known butyrate producers, microbes that normally support intestinal barrier integrity and immune regulation. Changes in these bacteria might thus reflect or even contribute to gut barrier breach and systemic inflammation in pancreatitis.
Interestingly, some previously reported associations with microbes like Roseburia and Ruminococcus were partly replicated, but many earlier findings diverged, underscoring the complexity and variability of gut-disease links. The study’s main takeaway is that while certain microbial signatures appear tied to disease severity and complications, the field still needs carefully designed, longitudinal research to tease apart cause from consequence and to understand how shifts in our microbial partners may drive or reflect critical illness.
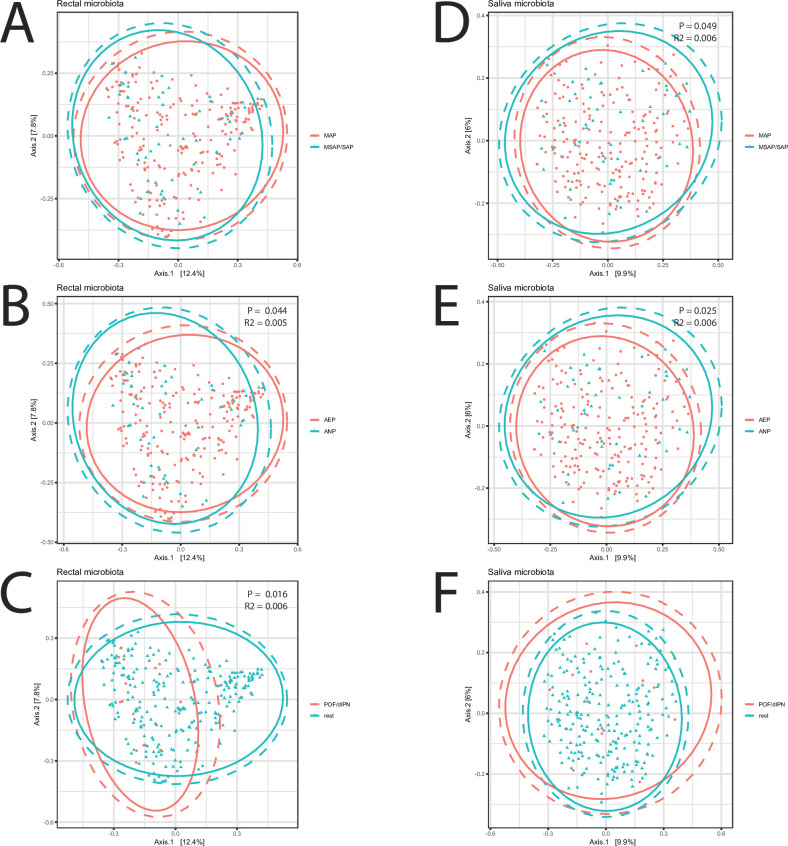
Figure 2. Beta diversity of bacterial communities. Principal coordinate analysis (PCoA) plot displaying Bray-Curtis distances of rectal (A–C) and saliva (D–F) microbiomes in groups of (A, D) MSAP and SAP versus MAP, (B, E) acute necrotising pancreatitis (ANP) versus acute oedematous pancreatitis (AOP), and (C, F) persistent organ failure (POF) and/or drained infected pancreatic necrosis (dIPN). Significance testing with PERMANOVA. MAP, mild acute pancreatitis; MSAP, moderate severe acute pancreatitis; SAP, severe acute pancreatitis.
Resilient but Altered: The Skin Microbiome in Critical Illness
When we think about the human microbiome, our gut often steals the spotlight but our skin hosts equally rich microbial ecosystems that help defend us against infection. In people critically ill in intensive care units (ICUs), that skin microbiome becomes unpredictable. This study investigated how daily antiseptic bathing with chlorhexidine or octenidine common practices meant to lower infection risk affects skin microbial communities compared with ordinary soap and water. What emerges is a nuanced story about resilience, dysbiosis, and the limits of our interventions in fragile microbiomes.
At baseline, ICU patients already showed a weakened skin microbiome compared with healthy individuals: the usual site-specific patterns of microbes across different body regions were lost, and bacteria more typical of other body niches appeared in unusual places, a signature of dysbiosis in critical illness. This disruption persisted through hospitalization and even after discharge, regardless of whether patients were bathed with antiseptics or simple soap and water.
Daily antiseptic bathing did reduce overall bacterial biomass on areas of the skin like the gluteal crease and armpit meaning fewer bacterial cells were present but it did not substantially change which bacterial groups were present or restore healthy microbial diversity. In other words, the broad community structure who’s there and in what balance remained largely intact despite repeated antiseptic exposure. Importantly, there was no meaningful increase in antimicrobial resistance genes in the microbiome, easing concerns that such practices might accelerate resistance evolution, at least in this context.
These findings suggest that the skin microbiome of ICU patients is both already perturbed by critical illness and remarkably resilient to daily antiseptic bathing at the community level. Rather than fundamentally reshaping skin microbial landscapes, antiseptics act more like temporary suppressants of bacterial load, leaving deeper ecological imbalances and their implications for infection risk and recovery largely unaltered.